BiographTA
Software & demoBiographTA Tokenizer
The rule-based tokenizer splits a text into sentences and makes sure that all tokens are separated from each other. To get a better performance on biomedical texts the tokenizer has been configured to accept brackets inside tokens (e.g.: 4-(2,6-Dimethoxyphenyl)butyl), abbreviations have been added (e.g.: al., Chem.) and a selection of bibliographic reference patterns have been added. BiographTA NeSp Scope Labeler & demo
We are preparing the release of the BiographTA Scope Labeler. A demo of the system can be accessed via this [ link ]. 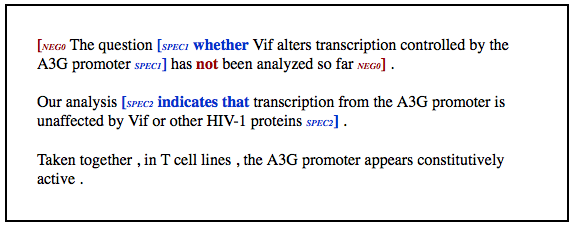
The system works in two phases. In the first phase, it finds negation and speculation cues using an SVM classifier. In the second phase, it finds the scope of the cues identified in the first phase. This is done with a memory-based system that relies on information from syntactic dependencies. The system has been trained on the data from the CoNLL Shared Task 2010 for speculation and on the BioScope corpus for negation. An example file with the output of the system can be found here. The system is based on the scope labeler described in: R. Morante, V. van Asch Vincent, W. Daelemans (2010) Memory-based resolution of insentence scopes of hedge cues. Proceedings of the Fourteenth Conference on Computational Natural Language Learning (CoNLL): Shared Task - CISB 978-1-932432-84-8 - Uppsala, Association for Computational Linguistics, 2010, p. 40-47. [PDF] NeSp in the BioNLP Shared Task 2011
The tool has been used to process the corpora from the BioNLP Shared Task 2011. The processed files can be found here. |


